23andMe co-founder Linda Avey commented on a previous post ("Identifying half-siblings by genetic tests"), pointing out that 23andMe provides images that illustrate full-siblings and half-siblings. In fact, they will represent any relationship pictorially. If you have an account there, you can compare your genome to another, with regions of half-identity and full-identity marked differently.
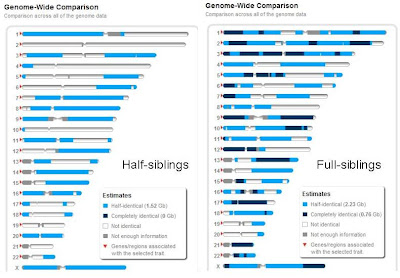
This pair of images shows regions of half-identity (light blue), complete identity (black)and no identity (white). The comparison on the left shows two women who share a father but have different mothers. Since they both have their father's complete X chromosome, they are half-identical across this entire chromosome. The rest of the genome is an even mix of regions that are half identical and regions that show no identity. On the right, you see a 1:2:1 ratio of no identity, half-identity and full identity. The most telling difference between these comparisons is the complete lack of regions with full identity when half-siblings are compared. If you subscribe to 23andMe, you can make similar images between any pair of shared genomes using their family inheritance genome view tool, at https://www.23andme.com/you/inheritance/.
Andrea Badger, who sent me these specific images, also collected data on the fraction of the genome identical by descent when different types of relative are compared. She found that full siblings shared between 41% and 55% of their genomes (16 comparisons) while half siblings shared between 21.7 and 31.4% of their genomes (four comparisons). The expected fractions are 50% and 25%, but there is some spread.
This is why having lots of markers is important. When the whole genome is analyzed, the difference is clear. With a smaller number of markers, statistical fluctuations are more likely to confuse the issue. 23andMe now uses over 1 million markers. Family Tree DNA uses fewer markers and charges more, but they use plenty of markers (hundreds of thousands) and have a greater focus on genealogy. I am aware of other services that charge almost as much but test only a few markers (less than 100). If you are shopping for a test, you can find a lot of information at dna-forums.
Thanks, Andrea!